About
The database
miRandola is a manually curated database containing information on extracellular and circulating non-coding RNAs. The first version of the database was published in Plos One in 2012 and it consisted of microRNA data extracted from public available papers in PubMed. The aim of miRandola is to collect all the information about non-coding RNAs that circulate in the blood stream and other body fluids. Non-coding RNAs play an important role in the regulation of various biological processes. They are frequently dysregulated in cancer, cardiovascular diseases and many other diseases and have shown promise as tissue-based markers for classification and prognostication. Extracellular non-coding RNAs in serum, plasma, saliva, urine and other body fluids have recently been shown to be associated with various pathological conditions. Non-coding RNAs circulate in the bloodstream in a highly stable, extracellular form, thus they may be used as blood-based biomarkers. The long term aim of miRandola is to become a comprehensive database for non-invasive biomarkers. We are working for new updates that will include other biomarkers such as extracellular DNA and circulating tumor cells. Users are welcome to download the database. The following table shows the comparison between the first published version of miRandola (2012) and the new version (2017):
Comparison between the first version of miRandola and the last version
| miRandola 2012 | miRandola 2017 | |
| *Papers | 89 | 314 |
|---|---|---|
| Entries | 2132 | 3283 |
| microRNAs | 581 | 1002 |
| Long non-coding RNAs | 0 | 12 |
| CircRNAs | 0 | 8 |
| Extracellular RNA forms | 4 | 7 |
| Drugs | 0 | 25 |
| Organisms | 1 | 14 |
| Sample types | 21 | 47 |
| Visualization tool | No | Yes |
| External data | ExoCarta | ExoCarta and Vesiclepedia |
| **Text-mining-assisted curation | No | Yes |
*See Articles in the database for more information
**See section Assisted Curation for more information
Assisted Curation
The first team of miRandola consisted of only one human biocurator. For this new version we increased the number of
biocurators involved in the project and moreover we introduced the Assisted Curation using a text mining approach.
It is important to underline that the new version of miRandola is still a manually curated database but we used
the text mining in order to search and prioritize the papers to curate through human curators. It is in fact clear that
the manual curation is a time consuming process and the approach we used will help our internal curators
to boost the update of the database at least two times per year (expected every 6 months).
The text mining dictionary included human non-coding RNAs [1], diseases [2], and keywords to indicate the RNA is present extracellularly and circulates.
Text mining was run on more than 26 million abstracts in PubMed using the tagger software developed in [3].
Pairs of these entities were scored by summing scores for all co-occurrences of the entities in the same sentence,
paragraph and document with decreasing weights. These scores were then normalized, and the geometric mean of the
RNA-disease and RNA-circulating scores was taken as the score for a triple.
Papers that contain all three types of entity were given the same score as the triple.
Papers were reviewed by human curators in order of decreasing score.
References
[1] Junge A, Refsgaard JC, Garde C, Pan X, Santos A, Alkan F, Anthon C, von Mering C, Workman CT, Jensen LJ, Gorodkin J. (2017). RAIN: RNA-protein Association and Interaction Networks. Database, baw167, 1–9. http://doi.org/10.1093/cercor/bhw393
[2] Pletscher-Frankild S, Pallejà A, Tsafou K, Binder JX, Jensen LJ. (2015). DISEASES: Text mining and data integration of disease-gene associations. Methods, 74, 83–89. http://doi.org/10.1016/j.ymeth.2014.11.020
[3] Pafilis E, Frankild SP, Fanini L, Faulwetter S, Pavloudi C, Vasileiadou A, Arvanitidis C, Jensen LJ. (2013). The SPECIES and ORGANISMS Resources for Fast and Accurate Identification of Taxonomic Names in Text. PLoS ONE, 8(6), 2–7. http://doi.org/10.1371/journal.pone.0065390
Search and query the database
In this new version of the database we introduced several ways to explore data in order to satisfy the need of different users. The user may be interested in browsing by data type (menu section):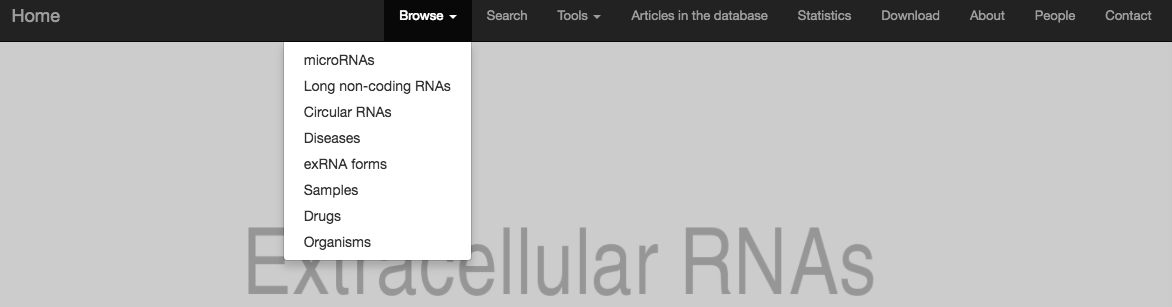 Then, by clicking on the specific data type, e.g. microRNAs, the user will explore the content with filtering and paging features:
Then, by clicking on the specific data type, e.g. microRNAs, the user will explore the content with filtering and paging features:
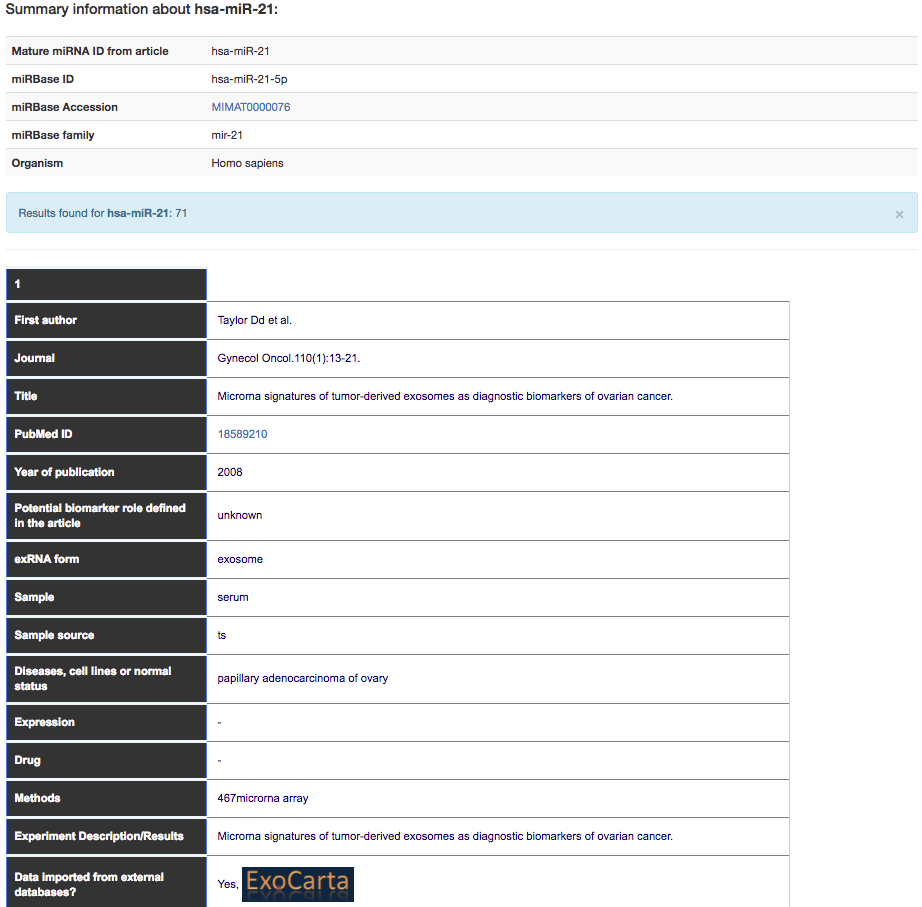
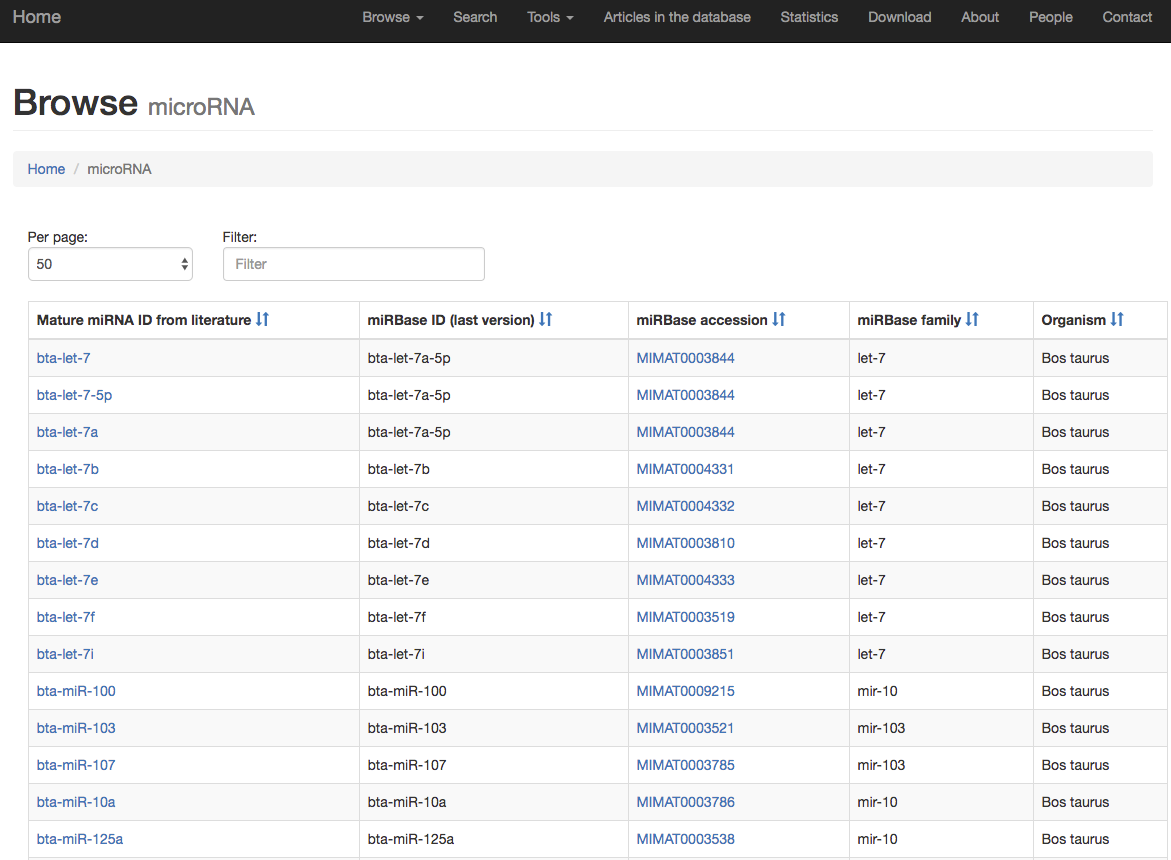 Clicking on the specific microRNA (in this example hsa-miR-21) the user will see more details about the entry in the database.
The details include information on the RNA of interest (e.g. miRBase accession number and ID), disease, sample type, extracellular RNA form (e.g. exosome), method to isolate and quantify the RNA, experiment description/results, organism, RNA expression, drug and information regarding the paper and other data.
To see this example click here.
Clicking on the specific microRNA (in this example hsa-miR-21) the user will see more details about the entry in the database.
The details include information on the RNA of interest (e.g. miRBase accession number and ID), disease, sample type, extracellular RNA form (e.g. exosome), method to isolate and quantify the RNA, experiment description/results, organism, RNA expression, drug and information regarding the paper and other data.
To see this example click here.
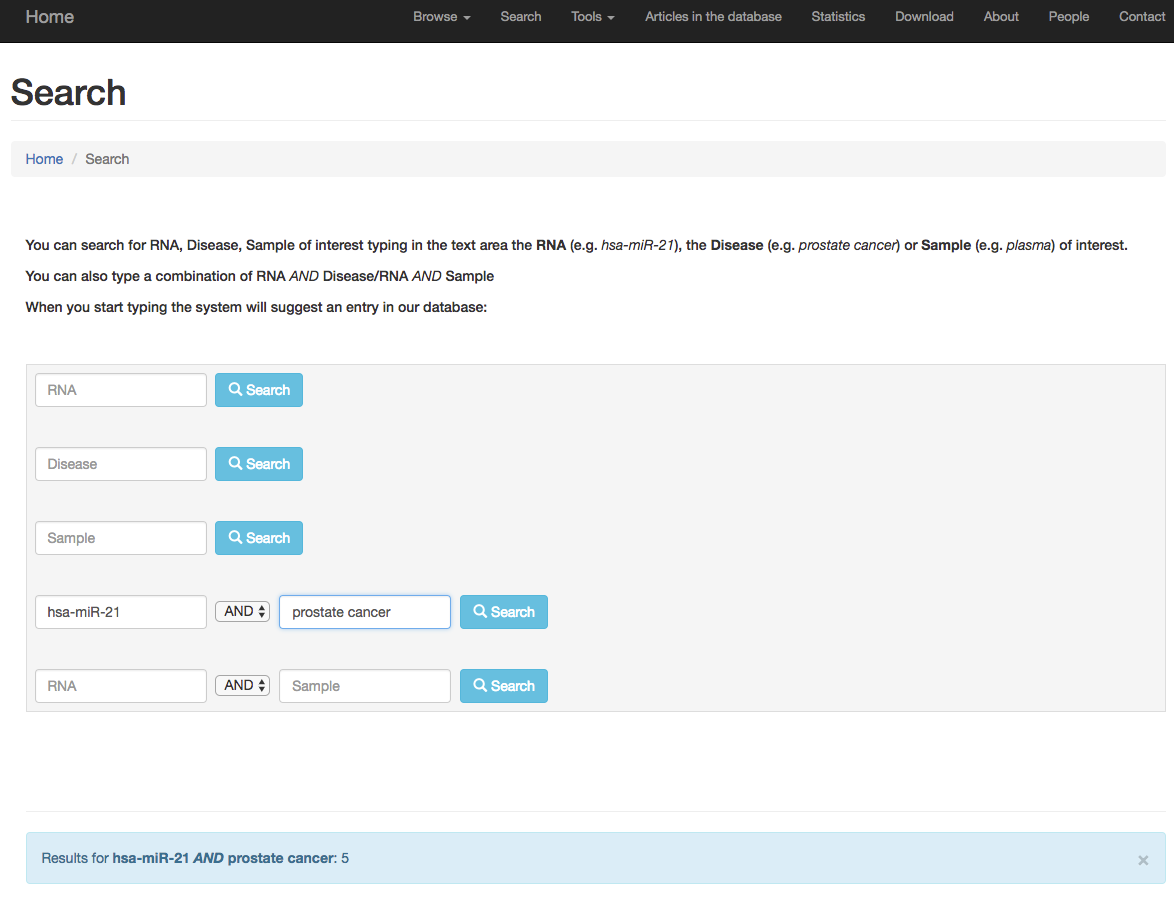
 The user may also be interested to search for specific RNA, disease or sample instead of browse the database. This is possible, using the
Search page (menu section). It is also possible to combine two terms of interest such as hsa-miR-21 and prostate cancer.
The user may also be interested to search for specific RNA, disease or sample instead of browse the database. This is possible, using the
Search page (menu section). It is also possible to combine two terms of interest such as hsa-miR-21 and prostate cancer.